| HOME | INSTRUMENTATION | NMR TIME | USERS & RESEARCH |
INDUSTRIAL AFFILIATES |
NEWS & LINKS |
Publications:
Lukavsky, PJ; Kim, I; Otto, GA; Puglisi, JD (2003) "Structure of HCV IRES domain II
determined by NMR" Nature Struct. Biol., Advance Online Publication
Li, Q; Khosla, C; Puglisi, JD; Liu, CW (2003) "Solution structure and backbone
dynamics of the holo form of the frenolicin acyl carrier protein" BIOCHEMISTRY,
42 4648-4657.
Lynch, SR; Gonzalez, RL; Puglisi, JD (2003) "Comparison of x-ray crystal structure of
the 30S subunit-antibiotic complex with NMR structure of decoding site oligonucleotide-paromomycin
complex" STRUCTURE, 11 43-53.
Kim, I., Lukavsky, P.J., and Puglisi, J.D. (2002) "NMR Study of 100 kDa HCV
IRES RNA Using Segmental Isotope Labeling" J. Am. Chem. Soc.,
124, 9338-39.
Lukavsky P.J., Puglisi J.D. (2001) "RNAPack: An integrated NMR approach to RNA structure determination" Methods, 25, 316-332.
Blanchard, S.C., and Puglisi, J.D. (2001) "Solution structure of the A loop
of 23S ribosomal RNA. Solution structure of the A loop of 23S ribosomal
RNA." PNAS, 98, 3720-21.
Lynch S.R., Puglisi J.D. (2001) "Structural origins of aminoglycoside specificity
for prokaryotic ribosomes" J. Mol. Biol., 306, 1037-1058.
Lynch S.R., Puglisi J.D. (2001) "Structure of a eukaryotic decoding region
A-site RNA" J. Mol. Biol., 306, 1023-1035.
(Details Below)
Lukavsky, P.J., Otto, G.A., Lancaster, A.M., Sarnow P. and Puglisi,
J.D. (2000) "Structures of two RNA domains essential for hepatitis C virus
internal ribosome entry site function" Nature Struct. Biol.,
7, 1105-1110.
Lynch, SR; Puglisi, JD (2000) "Application of residual dipolar coupling
measurements to identify conformational changes in RNA induced by antibiotics"
JOURNAL OF THE AMERICAN CHEMICAL SOCIETY, 122 7853-7854.
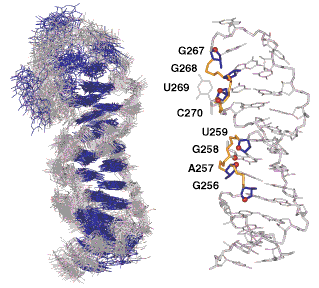
Details: Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function Lukavsky, P.J., Otto, G.A., Lancaster, A.M., Sarnow P. and Puglisi, J.D. (2000) Nature Struct. Biol. 7, 1105-1110. Abstract: Translation of the hepatitis C virus (HCV) polyprotein is initiated at an internal ribosome entry site (IRES) element in the 5' untranslated region of HCV RNA. The HCV IRES element interacts directly with the 40S subunit, and biochemical experiments have implicated RNA elements near the AUG start codon as required for IRES–40S subunit complex formation. The data we present here show that two RNA stem loops, domains IIId and IIIe, are involved in IRES–40S subunit interaction. The structures of the two RNA domains were solved by NMR spectroscopy and reveal structural features that may explain their role in IRES function.
|
| Bio-X | Structural Biology | School of Medicine | Stanford University |