| HOME | INSTRUMENTATION | NMR TIME | USERS & RESEARCH |
INDUSTRIAL AFFILIATES |
NEWS & LINKS |
Publications: (Details Below)
Hanick NA, Rickert M, Varani L, Bankovich AJ, Cochran JR, Kim DM, Surh CD, Garcia KC (2007) "Elucidation of the interleukin-15 binding site on its alpha receptor by NMR" Biochemistry 46 9453-61.
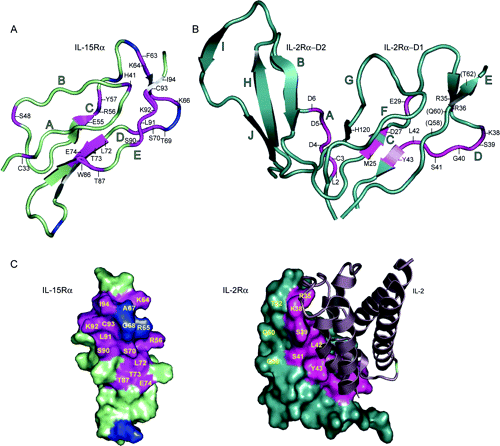
Details: Elucidation of the interleukin-15 binding site on its alpha receptor by NMR Hanick NA, Rickert M, Varani L, Bankovich AJ, Cochran JR, Kim DM, Surh CD, Garcia KC (2007) Biochemistry 46 9453-61. Abstract: The cytokine interleukin-15 (IL-15) signals through the formation of a quaternary receptor complex composed of an IL-15-specific alpha receptor, together with beta and gammac receptors that are shared with interleukin-2 (IL-2). The initiating step in the formation of this signaling complex is the interaction between IL-15 and IL-15Ralpha, which is a single sushi domain bearing strong structural homology to one of the two sushi domains of IL-2Ralpha. The crystal structure of the IL2-Ralpha/IL-2 complex has been determined, however little is known about the analogous IL-15Ralpha/IL-15 binding interaction. Here we show that recombinant IL-15 can be overexpressed as a stable complex in the presence of its high affinity receptor, IL-15Ralpha. We find that this complex is 10-fold more active than IL-15 alone in stimulating proliferation and survival of memory phenotype CD8 T cells. To probe the ligand/receptor interface, we used solution NMR to map chemical shifts on 15N-labeled IL-15Ralpha in complex with unlabeled IL-15. Our results predict that the binding surface on IL-15Ralpha involves strands C and D, similar to IL-2Ralpha. The interface, as predicted here, leaves open the possibility of trans-presentation of IL-15 by IL-15Ralpha on an opposing cell.
|
| Bio-X | Structural Biology | School of Medicine | Stanford University |